All AbMole products are for research use only, cannot be used for human consumption.
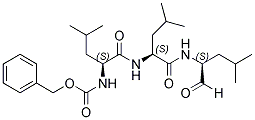
MG132 is a potent cell-permeable inhibitor of proteasome and calpain with IC50 of 100 nM and 1.2 μM respectively. MG132 inhibited the growth of HeLa cells via inducing the cell cycle arrest as well as triggering apoptosis. MG132 also inhibits NF-κB activation with an IC50 of 3 µM and prevents β-secretase cleavage. MG132 induces cell apoptosis through formation of reactive oxygen species or the upregulation and downregulation of these factors, which is ultimately dependent upon the activation of the caspase family of cysteine proteases.
In vitro, it induces neurite outgrowth in PC12 cells and has anticancer properties. Besides its apoptotic effect alone, MG132 also enhanced the antiglioma effect of the chemotherapeutics cisplatin, taxol and doxorubicin in C6 and U138MG cells, indicating an adjuvant/chemosensitizer potential.

J Cancer. 2025 Jan 01;16(2):417-429.
TYMS Enhances Colorectal Cell Antioxidant Capacity Via the KEAP1-NRF2 Pathway to Resist Ferroptosis
MG132 purchased from AbMole

Biochem Biophys Res Commun. 2025 Jan;742:151150.
O-GlcNAcylation of progranulin promotes hepatocellular carcinoma proliferation
MG132 purchased from AbMole

Adv Sci (Weinh). 2024 Jan;e2207435.
Defective Homologous Recombination Repair By Up-Regulating Lnc-HZ10/Ahr Loop in Human Trophoblast Cells Induced Miscarriage
MG132 purchased from AbMole

Autophagy. 2024 Jun 6.
The novel lnc-HZ12 suppresses autophagy degradation of BBC3 by preventing its interactions with HSPA8 to induce trophoblast cell apoptosis
MG132 purchased from AbMole

Environ Int. 2024 Sep;191:108975.
BaP/BPDE suppresses homologous recombination repair in human trophoblast cells to induce miscarriage: The roles of lnc-HZ08
MG132 purchased from AbMole

Int J Biol Sci. 2024 Aug 05.
The Antitumor and Sorafenib-resistant Reversal Effects of Ursolic Acid on Hepatocellular Carcinoma via Targeting ING5
MG132 purchased from AbMole

Int J Biol Macromol. 2024 Oct 30.
Circular RNA circWBSCR22 facilitates colorectal cancer metastasis by enhancing CHD4's protein stability
MG132 purchased from AbMole

Plant Physiol. 2024 Jan ;10:kiae011..
Transcription factor PbNAC71 regulates xylem and vessel development to control plant height
MG132 purchased from AbMole

Orphanet J Rare Dis. 2024 Nov 25;19(1):435.
Novel biallelic variants in IREB2 cause an early-onset neurodegenerative disorder in a Chinese pedigree
MG132 purchased from AbMole

Vet Microbiol. 2024 Jun 6;295:110150.
JEV infection leads to dysfunction of lysosome by downregulating the expression of LAMP1 and LAMP2
MG132 purchased from AbMole

biorxiv. 2024 Mar 25;14(7):999.
LncRNA-Snhg3 Aggravates Hepatic Steatosis by Regulating PPARγ SND1/H3K27me3
MG132 purchased from AbMole

bioRxiv. 2024 Aug 10.
LncRNA Snhg3 Aggravates Hepatic Steatosis via PPARgamma Signaling
MG132 purchased from AbMole

J Inflamm Res. 2023 Feb 27;16:861-877.
Neutrophil Extracellular Traps Induce Alveolar Macrophage Pyroptosis by Regulating NLRP3 Deubiquitination, Aggravating the Development of Septic Lung Injury
MG132 purchased from AbMole

Cell Death Differ. 2023 Oct;30(10):2213-2230.
LncRNA BCAN-AS1 stabilizes c-Myc via N6-methyladenosine-mediated binding with SNIP1 to promote pancreatic cancer
MG132 purchased from AbMole

Cancer Lett. 2023 Jul 28;567:216262.
The NF-Y splicing signature controls hybrid EMT and ECM-related pathways to promote aggressiveness of colon cancer
MG132 purchased from AbMole

J Transl Med. 2023 May 5;21(1):303.
SYTL2 promotes metastasis of prostate cancer cells by enhancing FSCN1-mediated pseudopodia formation and invasion
MG132 purchased from AbMole

Plant Physiol. 2023 Feb 12;191(2):1052-1065.
Calcyclin-binding protein-promoted degradation of MdFRUCTOKINASE2 regulates sugar homeostasis in apple
MG132 purchased from AbMole

BMC Biol. 2023 Apr 7;21(1):75.
1, 6-Hexanediol regulates angiogenesis via suppression of cyclin A1-mediated endothelial function
MG132 purchased from AbMole

Cell Death Discov. 2023 Apr 26.
Ammonium Tetrathiomolybdate Relieves Oxidative Stress in Cisplatin-induced Acute Kidney Injury via NRF2 Signaling Pathway
MG132 purchased from AbMole

FASEB J. 2023 May;37(5):e22931.
The novel peptide PFAP1 promotes primordial follicle activation by binding to MCM5
MG132 purchased from AbMole

World J Gastroenterol. 2023 Sep 21;29(35): 5104-5124.
Regenerating gene 4 promotes chemoresistance of colorectal cancer by affecting lipid droplet synthesis and assembly
MG132 purchased from AbMole

Cell Death Differ. 2022 Jul;29(7):1395-1408.
SARS-CoV-2 membrane protein causes the mitochondrial apoptosis and pulmonary edema via targeting BOK
MG132 purchased from AbMole

Ecotoxicol Environ Saf. 2022 Jun 1;237:113564.
Environmental BPDE induced human trophoblast cell apoptosis by up-regulating lnc-HZ01/p53 positive feedback loop
MG132 purchased from AbMole

Virol Sin. 2022 Aug 16;S1995-820X(22)00143-2.
CK1α upregulates the IFNAR1 expression to prompt the anti-HBV effect of type I IFN in hepatoma carcinoma cells
MG132 purchased from AbMole

Cell Biol Toxicol. 2022 Apr;38(2):291-310.
Lnc-HZ08 regulates BPDE-induced trophoblast cell dysfunctions by promoting PI3K ubiquitin degradation and is associated with miscarriage
MG132 purchased from AbMole

Bioengineered. 2022 Apr;13(4):8000-8012.
UBE2B promotes ovarian cancer growth via promoting RAD18 mediated ZMYM2 monoubiquitination and stabilization
MG132 purchased from AbMole

J Cell Mol Med. 2022 May;26(9):2646-2657.
Venetoclax enhances DNA damage induced by XPO1 inhibitors: A novel mechanism underlying the synergistic antileukaemic effect in acute myeloid leukaemia
MG132 purchased from AbMole

Exp Ther Med. 2022 Sep 23;24(5):687.
MG132 protects against lung injury following brain death in rats
MG132 purchased from AbMole

Patent. CN114053270A 2022 Feb 18.
Patent. CN114053270A
MG132 purchased from AbMole

Adv Sci (Weinh). 2021 Jan 6;8(4):2003205.
Inhibition Lysosomal Degradation of Clusterin by Protein Kinase D3 Promotes Triple-Negative Breast Cancer Tumor Growth
MG132 purchased from AbMole

Cell Death Dis. 2021 Aug 21;12(9):803.
USP14 maintains HIF1-α stabilization via its deubiquitination activity in hepatocellular carcinoma
MG132 purchased from AbMole

Cell Death Dis. 2021 Oct 16;12(11):955.
Essential role of zyxin in platelet biogenesis and glycoprotein Ib-IX surface expression
MG132 purchased from AbMole

PLoS Pathog. 2021 Feb;17(2):e100899.
CVB3 VP1 interacts with MAT1 to inhibit cell proliferation by interfering with Cdk-activating kinase complex activity in CVB3-induced acute
MG132 purchased from AbMole

Int J Mol Sci. 2021 Aug 16;22(16):8779.
Autophagy Mediates the Degradation of Plant ESCRT Component FREE1 in Response to Iron Deficiency
MG132 purchased from AbMole

J Cancer. 2021 Oct 17;12(23):7041-7051.
N6-methyladenosine regulates ATM expression and downstream signaling
MG132 purchased from AbMole

Oncogene. 2020 Apr;39(15):3163-3178.
The EGFR-ZNF263 Signaling Axis Silences SIX3 in Glioblastoma Epigenetically
MG132 purchased from AbMole

New Phytol. 2020 Jan;225(1):250-267.
Phospho-mutant activity assays provide evidence for alternative phospho-regulation pathways of the transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR.
MG132 purchased from AbMole

Mol Ther Nucleic Acids. 2020 Mar 6;19:1198-1208.
lncRNA RMST Suppressed GBM Cell Mitophagy through Enhancing FUS SUMOylation
MG132 purchased from AbMole

Mol Ther Nucleic Acids. 2020 Mar 6;19:1198-1208.
lncRNA RMST Suppressed GBM Cell Mitophagy through Enhancing FUS SUMOylation.
MG132 purchased from AbMole

Aging (Albany NY). 2020 Jul 17;12(14):14699-14717.
Acetylation-mediated degradation of HSD17B4 regulates the progression of prostate cancer
MG132 purchased from AbMole

Cell Microbiol. 2020 Mar;22(3):e13148.
Hepatitis B virus X protein modulates upregulation of DHX9 to promote viral DNA replication.
MG132 purchased from AbMole

Front Cell Infect Microbiol. 2020 Apr 16;10:158.
Difluoromethylornithine, a Decarboxylase 1 Inhibitor, Suppresses Hepatitis B Virus Replication by Reducing HBc Protein Levels
MG132 purchased from AbMole

Oncology Letters. 2019 Feb 8.
Inhibition of the PI3K/AKT signaling pathway sensitizes diffuse large B‑cell lymphoma cells to treatment with proteasome inhibitors via suppression of BAG3.
MG132 purchased from AbMole

Curr Biol. 2018 Aug 20;28(16):2616-2623.e5.
Chloroplast Biogenesis Controlled by DELLA-TOC159 Interaction in Early Plant Development.
MG132 purchased from AbMole
Oncotarget. 2018 Feb;9(12):10470-10482.
The hypoxia-responsive lncRNA NDRG-OT1 promotes NDRG1 degradation via ubiquitin-mediated proteolysis in breast cancer cells.
MG132 purchased from AbMole

Autophagy. 2017 Mar 4;13(3):538-553.
Acetylation targets HSD17B4 for degradation via the CMA pathway in response to estrone
MG132 purchased from AbMole

J Hematol Oncol. 2017 Jun 8;10(1):115.
SIX3, a tumor suppressor, inhibits astrocytoma tumorigenesis by transcriptional repression of AURKA/B
MG132 purchased from AbMole

Eur Respir J. 2015 Jun;1590-602.
Deleterious impact of Pseudomonas aeruginosa on cystic fibrosis transmembrane conductance regulator function and rescue in airway epithelial cells.
MG132 purchased from AbMole
| Cell Experiment | |
|---|---|
| Cell lines | Lung cancer cell lines A549 and H1299 |
| Preparation method | Cell viability assay. Cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells were seeded in 96-well plates at a density of 2.5x103/well 1 day prior to treatment. Then, cells were treated with MG132 or/and irradiation. After treatment, 20 µl of 5 mg/ml MTT solution was added into each well and incubated for 4 h. After the supernatant was removed, 100 µl of DMSO was added, and then placed in a microplate reader to measure OD value. Cell viability rate (vR) was calculated according to the following formula: vR = (OD in observed group/OD in 0 Gy group) x 100%. All assays were repeated 3 times in quintuplicate. |
| Concentrations | 200 nM |
| Incubation time | 6h |
| Animal Experiment | |
|---|---|
| Animal models | Male mdx (C57BL/10ScSn DMD mdx) mice |
| Formulation | Dissolved in DMSO, and diluted in PBS |
| Dosages | ~10 μg/kg/day |
| Administration | Injection |
| Molecular Weight | 475.62 |
| Formula | C26H41N3O5 |
| CAS Number | 133407-82-6 |
| Solubility (25°C) | DMSO 80 mg/mL Ethanol 20 mg/mL |
| Storage |
Powder -20°C 3 years ; 4°C 2 years In solvent -80°C 6 months ; -20°C 1 month |
[2] Ashley J Chui, et al. Science. N-terminal degradation activates the NLRP1B inflammasome
| Related Proteasome Products |
|---|
| Proteasome inhibitor IX
Proteasome inhibitor IX (PS-IX; AM114) is a Chalcone derivative and a chymotrypsin-like activity of the 20S proteasome inhibitor with an IC50 value of ~1 μM. |
| NIC-0102
NIC-0102 is an orally potent proteasome inhibitor (pIC50=7.55) that specifically inhibits NLRP3 inflammasome activation.NIC-0102 showed potent anti-inflammatory effects in vivo in a DSS-induced ulcerative colitis model. In addition, NIC-0102 inhibited the production of pro-IL-1β. |
| Ac-WLA-AMC
Ac-WLA-AMC is a specific 20S constitutive proteasome β5 fluorogenic substrate. |
| Ac-Nle-Pro-Nle-Asp-AMC
Ac-Nle-Pro-Nle-Asp-AMC is a specific substrate for 26S proteasome. |
| PR-39
PR-39, a natural proline- and arginine-rich antibacterial peptide, is a noncompetitive, reversible and allosteric proteasome inhibitor. |
All AbMole products are for research use only, cannot be used for human consumption or veterinary use. We do not provide products or services to individuals. Please comply with the intended use and do not use AbMole products for any other purpose.


Products are for research use only. Not for human use. We do not sell to patients.
© Copyright 2010-2024 AbMole BioScience. All Rights Reserved.
